- VSert.com/
- Posts/
- DataCurator.jl: Multi-threaded reproducible data curation with recipe based templates using R, Python, or Julia/
DataCurator.jl: Multi-threaded reproducible data curation with recipe based templates using R, Python, or Julia
Table of Contents
Reproducible scientific computing #
In scientific computing, it can be difficult to achieve reproducibility due to a number of factors:
- dependencies
- practitioners cannot change existing code or invoke it
- code in multiple languages needs to be combined
- hard to rerun poorly documented scripts
- parallelism is hard to achieve for non-experts
In this project, we set out to address the above challenges Code and paper are available online.
Note, images are sourced from this paper, to which I hold author copyright.
Architecture #
The idea is straightforward, describe in plain English how complex heterogeneous data should be organized, give this recipe to DC, which then converts the recipe to an executable template, and executes it.

Achieving parallellism #
Writing reproducible parallel code is hard, as it requires careful decomposition of interdependent tasks. DataCurator instead parallelizes where possible, but creating distinct execution threads, and deploying threading (shared memory) parallelism where possible. No explicit locking is done, avoiding costly contention. Instead, thread-local datastructures ensure that data is racefree until aggregation. This approach is not perfect though, as DC does not use the runtime information to tune its threading or scheduling policies.
Code free templates #
While DC has an API in Julia, it takes in code-free templates written in TOML.
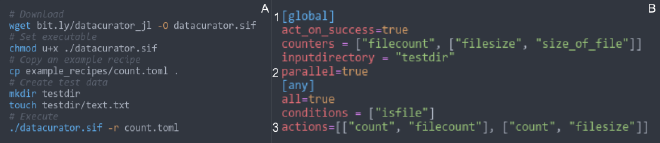
Combining R, Julia, and Python #
Combining code bases from different languages is difficult. Our approach is instead to implement DC in Julia, and use the outstanding interoperability APIs offered by RCall.jl and PyCall.jl) The result is that DC can reuse functions from Python, Julia, or R, as long as they are locally installed.
Typesafe dispatch #
Datacurator decodes recipes by decomposing them into functional components, then looking up those functions in its current and imported namespaces.
At inference, the functions are resolved by Julia by matching their type.
Test driven acceleration #
DC is tested thoroughly, but leverages tests for more than simply quality assurance. Because each new feature is accompanied by a test, DC’s core features are all tested. Regressions are resolved by adding test cases. This can be leveraged then by using Julia’s pre-compilation, as we can tell Julia’s profiler to watch execution during the test cases, then precompile any executed functions.
As a result, any tested feature is running at compile speeds.
A full scientific computing workflow #
The above discussion the brings us to the following flowchart that lays out the different steps involved in scientific computing.

Future work #
While the emergence of LLMs made scientific computing easier for experts, their ability to hallucinate non-existing function calls or at times generate non-sensical, but correct sounding code, only highlights the importance of frameworks that enforce correctness over hype. This is not to say that generative solutions do not have their place, but that their usage is to be supervised by experts.
In future work the work scheduling of DC can be executed more intelligently, distributing it based on the expected load.
Publication #
DataCurator was published in the following work:
- Ben Cardoen, Hanene Ben Yedder, Sieun Lee, Ivan Robert Nabi, and Ghassan Hamarneh. DataCurator.jl: Efficient, portable, and reproducible validation, curation, and transformation of large heterogeneous datasets using human-readable recipes compiled into machine verifiable templates. BioInformatics Advances, 3(1-5):dx.doi.org/10.1093/bioadv/vbad068, 2023.